Evolution of rabbit haemorrhagic disease virus in New Zealand

Image - John Hunt
Since its illegal introduction in 1997, rabbit haemorrhagic disease (RHD) virus has spread throughout New Zealand and greatly reduced rabbit populations in many regions.
However, rabbit numbers in some areas are now recovering as an increasing proportion of rabbits have developed protective antibodies against the disease. This allows them to survive any subsequent exposure to the RHD virus. Rabbits can acquire the protective antibodies following exposure to the RHD virus early in life or possibly from infection by a closely-related but benign form of rabbit calicivirus that was present in New Zealand rabbits before the RHD virus was introduced. Janine Duckworth and the Rabbit Biocontrol Initiative team have been working to identify ways to maintain and improve the ability of the RHD virus to kill rabbits. Two key questions being addressed by the team are whether the RHD virus has changed since its release, such that different strains now exist in New Zealand, and, if so, do the strains differ in their ability to kill rabbits.
With assistance from local farmers, rabbit control contractors and land users, Janine and the Rabbit Biocontrol Initiative have collected 24 wild New Zealand field strains of RHD virus (Fig. 1) and genetically tested them. Such analysis shows that RHD virus has evolved (Fig. 2), is now genetically distinct from the original virus (Czech V351 strain) released in New Zealand, and is also different to wild strains of RHD found in Australia.
Eight of the wild New Zealand RHD strains were selected for testing in domestic rabbits to see if genetic changes in the virus resulted in changes in its pathogenicity. The team’s aim was to identify New Zealand RHD strains that killed quickly and humanely. Results indicate that all the strains of RHD virus tested were lethal to rabbits (Table) and the overall kill rate was 97%. All but two of the New Zealand strains were similar in pathogenicity to the originally released Czech V351 strain. One exception was isolate 64 Otago Alexandra, which took significantly longer to kill (125.6 ± 1.7 hours from exposure to death), with a longer period of fever (57.5 ± 7.2 hours), indicating poor efficacy and animal welfare impacts. For those reasons the 64 Otago Alexandra strain was not selected for further evaluation. In contrast, strain 95 Mackenzie Basin caused death significantly faster (39.9 ± 1.7 hours) than the Czech V351 strain (74.3 ± 16.0 hours), (Table) and animal welfare impacts were acceptable. This indicates that strain 95 Mackenzie Basin is a highly pathogenic strain of RHD virus and worthy of further investigation. The strain is currently being assessed for its ability to kill wild rabbits, including those with antibodies against benign rabbit calicivirus.
Concurrently, molecular techniques were used to confirm the presence and identification of benign rabbit caliciviruses present in New Zealand. Serum antibody levels measured by Leila Nicholson (PhD student) showed that benign rabbit calicivirus is widely spread throughout the country, and recently Leila isolated and identified the first New Zealand strain of benign rabbit calicivirus. Janine and her team are now determining the extent to which prior exposure to benign rabbit calicivirus protects rabbits against subsequent RHD virus challenge, and quantifying its potential to impact on the effectiveness of RHD biocontrol.
In the future, and subject to gaining appropriate approval, high-virulence strains of New Zealand-sourced RHD virus, and best practice methods for their use to minimise any effect of pre-existing benign rabbit caliciviruses on RHD impacts, may be made available to land managers to maximise the benefits of rabbit control and as tools supporting long-term control of wild rabbits in New Zealand.
This work is funded by the Ministry for Primary Industry’s Sustainable Farming Fund (12/055) and the Australian Invasive Animal Co-operative Research Centre.
Janine Duckworth
Leila Nicholson (Landcare Research/University of Otago)
Katherine Trought
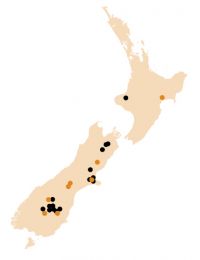
Fig. 1 Map showing sites in New Zealand where wild strains of RHD virus were collected. Orange dots indicate strains tested for pathogenicity.
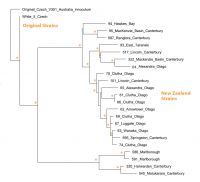
Fig. 2 New Zealand-sourced RHD virus strains clustered on phylogenetic analysis of gene sequences for the viral coat protein (VP60). Bootstrap probability values above 75% are indicated by asterisks at the nodes.
Table. Effect of RHD virus strain on kill rate, time from oral dosing (exposure) to death and time from onset of fever to death. Values within columns assigned different letters are significantly different (P<0.05).
