'Hotspots' of endemism (biodiversity found only in NZ)
Peter Heenan
Captain Cook’s first (1769) and second (1773-74) voyages to New Zealand produced the first scientific collections of New Zealand native plants, collected by expedition botanists who preserved specimens for the British Museum1. Subsequent explorers and ‘scientist-settlers’ collected more specimens from across New Zealand, with information about locality, date, taxa and other interesting details recorded on labels and in diaries. These early botanical collections now reside in New Zealand and offshore herbaria alongside more modern collections, with well over one million specimens of native flowering plants, ferns, conifers and cryptogams having been collected from New Zealand. Many of these specimens are truly unique to New Zealand, i.e. they are endemic species occurring nowhere but here (82% of our native plant species are endemic).
Since those early records, information on our native and endemic plants and where they grow has gradually increased. The accumulating information reveals that some localities have many endemic species or unique combinations of species.
With the advent of computerised databases, the locality information for many of these specimens could be captured electronically with geo-reference co-ordinates. Botanists have traditionally used geo-references to map individual species distributions, with aggregated species distributions being used to manually identify broad areas of endemism.
Next generation biodiversity analyses
The rapidly accumulating availability of electronic geo-referenced spatial records for herbaria around the world has led to international efforts to develop new statistically robust biodiversity analyses and reporting tools, such as Biodiverse. Biodiverse was developed specifically for spatial analyses of diversity using indices based on taxonomic, phylogenetic, trait and matrix-based (e.g. genetic distance) relationships, as well as related environmental and temporal variations. In a ‘proof-of-concept’ project, we assessed whether the free Biodiverse software and digitised herbarium records could be used to identify important areas of endemism for conservation prioritisation, planning and management and environmental reporting.
Vascular flora for the entire New Zealand archipelago covered
We used georeferenced spatial data covering the entire indigenous vascular flora ‒ the taxonomically diverse fern, conifer and flowering plant genera and species. The entire New Zealand archipelago of over 700 islands ‒ the mainland islands, inshore islands and the more distant offshore islands, such as the northern-most Kermadec and Three Kings Islands, the mid-latitude Chatham Islands, and the Sub Antarctic islands ‒ was included in the project.
We also developed a DNA phylogeny showing relationships of all of the New Zealand genera of ferns, conifers and flowering plants to enable phylogenetic (evolutionary ‘relatedness’) metrics to be analysed in conjunction with the spatial data. Undertaking spatial and phylogenetic analyses for the entire flora of an archipelago is unique; most previous studies have focused on genera and families that occur on continents.
A significant benefit of this integrated approach is that it produces specific results quantifying the area of endemism and qualifying the type of endemism, such as species- or generic-level endemism, phylogenetic and neo-or palaeo-endemism. This is the first study to demonstrate the type of endemism occurring in particular regions.
More detailed information about endemism
The results of where the endemism occurs are broadly consistent with earlier studies but are more accurate and detailed.
- All offshore islands have high species-level endemism
- On mainland New Zealand, species-level endemism occurs on Stewart Island, mountain areas of northern Southland, central Otago plateau uplands, the Arthur Range in Nelson, South Canterbury and inland and coastal Marlborough. In the North Island, it occurs only at the Surville Cliffs ‒ the northern-most tip of the country
- Genus-level and genus phylogenetic endemism predominantly occurs in the northern half of the North Island and northern offshore islands.
- Neo-endemism prevails in the South Island mountains, the result of recent and rapid species radiations in some plant lineages. In the northern North Island, palaeo-endemism prevails as the result of accumulated older plant lineages.
Maps highlighting the areas and types of endemism have been generated from the analyses (see graphs).
Significance of the results
This study has successfully used a range of spatial biodiversity metrics for the entire vascular flora of the New Zealand archipelago. It has revealed new centres of endemism and confirmed previously identified ‘hotspots’, but with more accuracy than before. Furthermore, it is the first qualitative assessment of the type of endemism.
The study also highlights the significant value of the Nationally Significant Collections and Databases for which Landcare Research acts as a custodian on behalf of all New Zealanders. Our Collections include the Allan Herbarium, the largest and most significant herbarium in New Zealand, as well as other national biological Collections. We have invested significant Core funding over recent years to ‘digitise’ information and images from these Collections and ensure this valuable digital information can be easily accessed from Landcare Research Databases.
This research will contribute significantly to future regional and national scales of conservation as many areas and types of endemism are often poorly protected in the Department of Conservation (DOC) estate. For example:
- 40% of the areas that are hotspots for endemic species occur within the DOC conservation estate
- 29% of hotspots for endemic genera are within the conservation estate but only 19% of areas with high phylogenetic endemism are included
- only 15% of areas with high neo- and palaeo-endemism are within the conservation estate
This means we have not been very successful in protecting some of the most important areas that contain the ‘evolutionary potential’ for the New Zealand flora. Many of the areas of endemism that occur in lowland and coastal areas are not adequately protected. The centres and types of endemism identified urgently need to be considered in conservation policy, planning and management.
Improved conservation planning and prioritisation
The new range of metrics will enable scientifically sound decisions for using limited conservation resources to target the most important areas. At the national scale, organisations such as DOC, QEII National Trust and the Nature Heritage Fund should be able to use our data for conservation planning and prioritisation to ensure that the most valuable areas of New Zealand biodiversity are protected and managed in perpetuity.
End user interest and adoption
The Ministry for the Environment (MfE) has expressed a strong interest in using the metrics on endemism for biodiversity reporting because they are statistically robust, can be applied locally and nationally and resolved to different spatial scales, and enable trends to be reported. MfE recognises the value of the genetic metrics in biodiversity reporting, and has commissioned DNA sequencing to ensure the DNA phylogeny is based entirely on sequences from New Zealand species.
DOC and QEII National Trust have expressed an interest in the metrics for the identification of significant natural areas and for spatial prioritisation of threatened ecosystems for conservation.
Key research collaborators
Data was derived from the New Zealand Virtual Herbarium (NZVH), a collaborative initiative supported by ten herbaria with significant collections of indigenous plants. Landcare Research’s Allan Herbarium is a significant contributor to NZVH, and is primarily supported by Core funding.
When adopted:
1Some of these specimens have been gifted back to New Zealand and are now housed in Landcare Research’s Allan Herbarium at Lincoln.
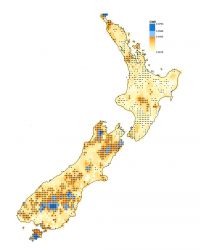
Map1: Species-level endemism for the top 1% (dark blue) and 5% (light blue) of cells. + represents cells with greater variation than expected from random; • represents cells with less variation than expected from random.
![<strong>Map 2:</strong> Cells with the top 5% of the combined species, genus and genus phylogenetic areas of endemism that occur in the DOC conservation estate (dark green) or in unprotected areas (tan). [Light green are the forested parts of New Zealand]](https://oldwww.landcareresearch.co.nz/__data/assets/image/0004/104575/varieties/thumb200.jpg)
Map 2: Cells with the top 5% of the combined species, genus and genus phylogenetic areas of endemism that occur in the DOC conservation estate (dark green) or in unprotected areas (tan). [Light green are the forested parts of New Zealand]
